Chapter 2 Taxonomic analysis
2.1 Update taxon names and clean up
Some MyCoPortal records will have outdated or invalid taxonomy; therefore, all taxon names need to be either updated or confirmed using currently accepted taxonomic classification (e.g., GBIF backbone taxonomy) prior to trait analyses. In the example below, records not identified to the species level are removed from the data set. This helps avoid taxonomy uncertain issues, which are described in more detail by Simpson and Schilling (2021).
After updating taxonomy, some records may now be classified outside of the taxonomic group that was originally queried. These records need to be removed if your analysis is to focus on the queried taxonomic group only.
Example code for updating taxon names and cleaning up data set:
library(fungarium)
#import sample data set
data(agaricales) #US agaricales records
#see Table 2.1 for preview of agaricales data set| id | institutionCode | scientificName | scientificNameAuthorship | recordedBy | year | eventDate | occurrenceRemarks | habitat | associatedTaxa | country | stateProvince | county | decimalLatitude | decimalLongitude | references | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 269864 | 9961160 | O | Galerina mniophila | (Lasch) Kühner | Gro Gulden | 1997 | 1997-11-15 | young Douglas fir forest | United States | Oregon | Benton | https://mycoportal.org/portal/collections/individual/index.php?occid=9961160 | ||||
| 313673 | 454144 | MICH | Mycena galopus | (Pers.) P. Kumm. | A. H. Smith | 1935 | 1935-11-30 | USA | California | Humboldt | 41.0595 | -124.1419 | https://mycoportal.org/portal/collections/individual/index.php?occid=454144 | |||
| 157092 | 4570238 | iNaturalist | Armillaria mellea | (Vahl) P. Kumm. | Cindy Trubovitz | 2017 | 2017-01-07 | <p>Growing from the base of an old log</p>; <a href=‘https://www.inaturalist.org/observations/5244247’ target=’_blank’ style=‘color: blue;’> Original observation #5244247 (iNaturalist)</a> | United States | California | 32.80975 | -117.1295 | https://mycoportal.org/portal/collections/individual/index.php?occid=4570238 | |||
| 281279 | 456630 | MICH | Hygrophorus olivaceoalbus | (Fr.) Fr. | R. L. Shaffer | 1956 | 1956-09-16 | USA | Idaho | Valley | 44.8753 | -115.9017 | https://mycoportal.org/portal/collections/individual/index.php?occid=456630 | |||
| 32558 | 2270252 | MICH | Cortinarius | (Pers.) Gray | S. J. Mazzer | 1970 | 1970-05-31 | [Collection is stored in Cortinarius indeterminate box] | USA | Michigan | Cheboygan | https://mycoportal.org/portal/collections/individual/index.php?occid=2270252 | ||||
| 522392 | 345316 | MICH | Crepidotus malachius var. phragmocystidiosus | Hesler & A.H. Sm. | A. H. Smith | 1959 | 1959-08-18 | Notes with collection North Amer. Sp. Crepidotus 1965 pg. 58. | On hardwood | USA | Michigan | Chippewa | https://mycoportal.org/portal/collections/individual/index.php?occid=345316 | |||
| 234305 | 526338 | OSC | Cortinarius montanus | Kauffman | Jim Trappe | 1976 | 1976-09-08 | Old-growth Tsme | Old-growth Tsme | USA | Oregon | Lane | https://mycoportal.org/portal/collections/individual/index.php?occid=526338 | |||
| 356189 | 2287069 | DUKE | Schizophyllum commune | Fr. | D. Porter | 1998 | 1998-04-00 | United States | Georgia | Clarke | 33.95 | -83.383333 | https://mycoportal.org/portal/collections/individual/index.php?occid=2287069 | |||
| 237907 | 2357852 | WTU | Cortinarius rubicundulus | (Rea) A. Pearson | L. L. Norvell, SAR. | 1992 | 1992-11-17 | Rotten wood/ duff. Sequoia, Tan Oak. | U.S.A. | California | Mendocino | https://mycoportal.org/portal/collections/individual/index.php?occid=2357852 | ||||
| 140361 | 6920708 | iNaturalist | Amanita pachycolea | D.E. Stuntz | Taylor Bates | 2017 | 2017-11-13 | <a href=‘https://www.inaturalist.org/observations/8797347’ target=’_blank’ style=‘color: blue;’> Original observation #8797347 (iNaturalist)</a> | United States | Oregon | 44.665631 | -123.242596 | https://mycoportal.org/portal/collections/individual/index.php?occid=6920708 |
#update names and classification
data <- taxon_update(agaricales, species_only=TRUE, show_status=FALSE)
#remove records with taxon names that could not be updated or confirmed
data2 <- data[data$new_species!="",]
#remove records no longer classified in agaricales
data3 <- data2[data2$new_order=="Agaricales",] Note that taxon name updating and filtering was done prior to the taxonomic, geographic, and temporal analyses done by Simpson and Schilling (2021). This is not necessarily required for geographic and temporal analyses; however, you run the risk of including “bad taxa” (i.e. taxa that are not actually classified within your originally specified taxonomic group) in your analyses. Thus, it is recommended to update taxon names and filter before performing any downstream analyses.
2.2 Calculating trait enrichment factors
To make quantitative trait comparisons between taxa, trait-relevant records are first identified using find_trait and then trait enrichment factors are calculated using enrichment. These enrichment factors correspond to the number of records associated with a certain trait divided by the number of total records for a given taxon. The following example investigates fire-association (i.e., association with fire-affected habitat, hosts, or substrates) as an ecological trait.
Example code for finding trait records and calculating enrichment factors:
#string for finding fire-associated records
string1 <- "(?i)charred|burn(t|ed)|scorched|fire.?(killed|damaged|scarred)|killed.by.fire"
#string for removing records falsely identified as fire-associated
string2 <- "(?i)un.?burn(t|ed)"
#filter out records that do not contain any environmental metadata (optional)
#note that "host" is no longer present in new MyCoPortal data sets; see find_trait documentation
data4 <- data3[data3$occurrenceRemarks!=""|data3$associatedTaxa!=""|
data3$habitat!="",]
#find trait-relevant records
trait_data <- find_trait(data4,
pos_string=string1, neg_string=string2)
#get trait enrichment
trait_enrichment <- enrichment(all_rec=data, trait_rec=trait_data, coll_bias=TRUE, status_feed=FALSE)
#filter taxa based on collector bias (optional)
trait_enrichment <- trait_enrichment[trait_enrichment$max_bias<=0.75,]
#filter taxa based on total number of records (optional)
trait_enrichment <- trait_enrichment[trait_enrichment$freq>=5,]Note that there are various optional filtering steps that may be done before and after trait searching and enrichment factor calculation, all of which have the potential to improve the accuracy of enrichment values. In the example above, records that did not contain any environmental metadata were removed prior to trait searching. This helps remove potential false negative records (i.e., records that are in fact associated with the trait of interest but are not identified as such), which negatively impact the enrichment factor accuracy. After trait searching and enrichment factor calculation, there a variety of output fields that can be used for data set filtering.
The “freq” or “trait_freq” output fields correspond to the number of total records and the number of trait-relevant records, respectively, for each unique taxon. These can both be used to filter out taxa that have not been sampled thoroughly enough to calculate trustworthy enrichment values (enrichment values are listed under the “trait_ratio” output field; see enrichment documentation for more details about output fields). For example, a taxon with 50 total records will have a more trustworthy enrichment value than a taxon that has only 5 total records.
The “max_bias” or “coll_groups” output fields correspond to the maximum proportion of records collected by one group of associated collectors and the total number of associated collector groups, respectively. These fields can be used to filter out taxa that have been collected with a high degree of bias, and, thus, may not have trustworthy enrichment values. For example, a taxon with a “max_bias” value of 0.25 will likely have a more trustworthy enrichment value than a taxon with a value of 0.85. See Simpson and Schilling (2021) for more details about how enrichment values can be negatively impacted by collector bias.
2.3 Visualize trait enrichment factors
After trait enrichment factors have been calculated for each species, these values can been visualized within a cladogram using trait_clado. This function generates a cladogram based on the taxonomic classification of each species (up-to-date taxonomic classification is given in the output of taxon_update). Cladogram tip color corresponds to the enrichment value of each species, whereas node color corresponds to the enrichment value of each higher-level taxon in the cladogram. Other tree annotation options are available. See trait_clado documentation for more details.
Example code for making rectangular trait cladogram:
#load sample enrichment data set for global Agaricales records
data(agaricales_enrich)
#see Table 2.2 for preview of Agaricales enrichment data set| new_full_name | freq | trait_freq | trait_ratio | coll_blanks | blanks_bias | coll_blanks_t | blanks_bias_t | max_bias | coll_groups | max_bias_t | coll_groups_t | new_kingdom | new_phylum | new_class | new_order | new_family | new_genus | new_species | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1626 | Tricholoma colossus (Fr.) Quél. | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0.5000000 | 2 | 0 | 0 | Fungi | Basidiomycota | Agaricomycetes | Agaricales | Tricholomataceae | Tricholoma | Tricholoma colossus |
| 555 | Harmajaea harperi (Murrill) Dima & P.Alvarado | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1.0000000 | 1 | 0 | 0 | Fungi | Basidiomycota | Agaricomycetes | Agaricales | Pseudoclitocybaceae | Harmajaea | Harmajaea harperi |
| 2135 | Coprinopsis acuminata (Romagn.) Redhead, Vilgalys & Moncalvo | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0.4000000 | 4 | 0 | 0 | Fungi | Basidiomycota | Agaricomycetes | Agaricales | Psathyrellaceae | Coprinopsis | Coprinopsis acuminata |
| 2478 | Coprinopsis cinerea (Schaeff.) Redhead, Vilgalys & Moncalvo | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0.1111111 | 9 | 0 | 0 | Fungi | Basidiomycota | Agaricomycetes | Agaricales | Psathyrellaceae | Coprinopsis | Coprinopsis cinerea |
| 2342 | Amanita agglutinata (Berk. & M.A.Curtis) Lloyd | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0.2857143 | 6 | 0 | 0 | Fungi | Basidiomycota | Agaricomycetes | Agaricales | Amanitaceae | Amanita | Amanita agglutinata |
| 885 | Mycena subrubridisca (Murrill) Murrill | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1.0000000 | 1 | 0 | 0 | Fungi | Basidiomycota | Agaricomycetes | Agaricales | Mycenaceae | Mycena | Mycena subrubridisca |
| 2281 | Cortinarius porphyropus (Alb. & Schwein.) Fr. | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0.3333333 | 5 | 0 | 0 | Fungi | Basidiomycota | Agaricomycetes | Agaricales | Cortinariaceae | Cortinarius | Cortinarius porphyropus |
| 540 | Gymnopus dentatus Murrill | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1.0000000 | 1 | 0 | 0 | Fungi | Basidiomycota | Agaricomycetes | Agaricales | Omphalotaceae | Gymnopus | Gymnopus dentatus |
| 2450 | Melanoleuca dryophila Murrill | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0.1250000 | 8 | 0 | 0 | Fungi | Basidiomycota | Agaricomycetes | Agaricales | Tricholomataceae | Melanoleuca | Melanoleuca dryophila |
| 1013 | Psathyrella incondita A.H.Sm. | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1.0000000 | 1 | 0 | 0 | Fungi | Basidiomycota | Agaricomycetes | Agaricales | Psathyrellaceae | Psathyrella | Psathyrella incondita |
#filter out taxa with high collector bias (optional)
agaricales_enrich <- agaricales_enrich[agaricales_enrich$max_bias<=0.75,]
agaricales_enrich <- agaricales_enrich[agaricales_enrich$max_bias_t<=0.75,]
#filter out taxa with low total records ("freq")
agaricales_enrich <- agaricales_enrich[agaricales_enrich$freq>=3,]
#make cladogram
library(ggtree)
library(ggplot2)
trait_clado(data=agaricales_enrich, continuous="color",
ladderize=TRUE, layout="rectangular", size=1.2,
formula = ~new_order/new_family/new_genus/new_species,
filter="high-100-trait_ratio", node_all = TRUE)+
geom_tiplab(color = "black", hjust = 0, offset = 0.1,
size = 2.6, fontface = "italic") + #add species labels
geom_tippoint(shape=20,
aes(color=trait_ratio, size=trait_freq),
alpha=0.75)+#add tree tips
scale_color_gradientn(colours= c("cyan", "blue", "purple", "red", "orange"),
name = "Fire-associated records enrichment",
limits = c(0, round(max(agaricales_enrich$trait_ratio),2)),
guide = guide_colourbar(label.vjust = 0.6,
label.theme = element_text(size = 10,
colour = "black",
angle = 0),
title.position = "top",
nbin=100,
draw.ulim = FALSE,
draw.llim = FALSE,
barwidth = 15,
barheight = 0.5))+
scale_size(name = "Fire-associated records",
guide = guide_legend(keywidth = 2,
keyheight = 1,
label.position = "bottom",
label.vjust = 0.6,
label.theme = element_text(size = 10,
colour = "black",
angle = 0),
title.position = "top")) +
theme(plot.margin=margin(0,0,0,0),
legend.title = element_text(size = 10, margin = margin(0,0,0,0)),
legend.title.align = 0.5,
legend.position = "bottom",
legend.justification = "center",
legend.margin = margin(0,0,0,0),
plot.title = element_text(hjust = 0.5, margin=margin(0,0,0,0)))+
xlim(c(0,5))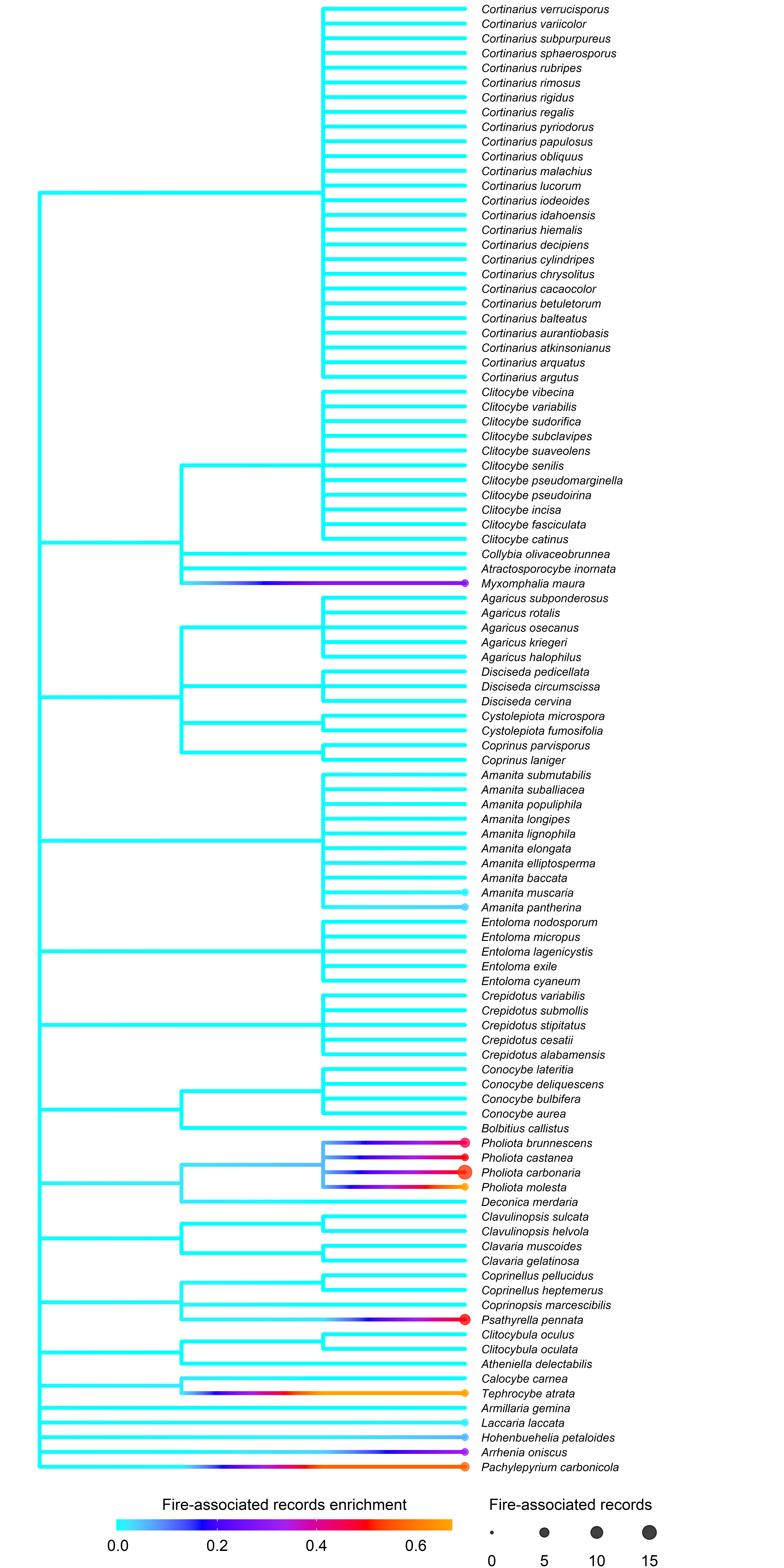
Figure 2.1: Cladogram showing fire-association enrichment for top 100 Agaricales species with the highest enrichment
Example code for making circular trait cladogram:
#load sample enrichment data set
data(agaricales_enrich)
#filter out taxa with high collector bias (optional)
agaricales_enrich <- agaricales_enrich[agaricales_enrich$max_bias<=0.75,]
agaricales_enrich <- agaricales_enrich[agaricales_enrich$max_bias_t<=0.75,]
#filter out taxa with low total records ("freq")
agaricales_enrich <- agaricales_enrich[agaricales_enrich$freq>=3,]
#make cladogram
library(ggtree)
library(ggplot2)
trait_clado(data=agaricales_enrich, continuous="color",
ladderize=TRUE, layout="circular", size=0.8,
formula = ~new_order/new_family/new_genus/new_species,
filter="high-300-trait_ratio", node_all = TRUE)+
geom_tiplab2(color = "black", hjust = 0, offset = 0.1,
size = 1, fontface = "italic") + #add species labels
geom_tippoint(shape=20,
aes(color=trait_ratio, size=trait_freq),
alpha=0.75)+#add tree tips
scale_color_gradientn(colours= c("cyan", "blue", "purple", "red", "orange"),
name = "Fire-associated records enrichment",
limits = c(0, round(max(agaricales_enrich$trait_ratio),2)),
guide = guide_colourbar(label.vjust = 0.6,
label.theme = element_text(size = 10,
colour = "black",
angle = 0),
title.position = "top",
nbin=100,
draw.ulim = FALSE,
draw.llim = FALSE,
barwidth = 15,
barheight = 0.5))+
scale_size(name = "Fire-associated records",
guide = guide_legend(keywidth = 2,
keyheight = 1,
label.position = "bottom",
label.vjust = 0.6,
label.theme = element_text(size = 10,
colour = "black",
angle = 0),
title.position = "top")) +
theme(plot.margin=margin(0,0,0,0),
legend.title = element_text(size = 10, margin = margin(0,0,0,0)),
legend.title.align = 0.5,
legend.position = "bottom",
legend.justification = "center",
legend.margin = margin(0,0,0,0),
plot.title = element_text(hjust = 0.5, margin=margin(0,0,0,0)))+
xlim(c(-1, 4))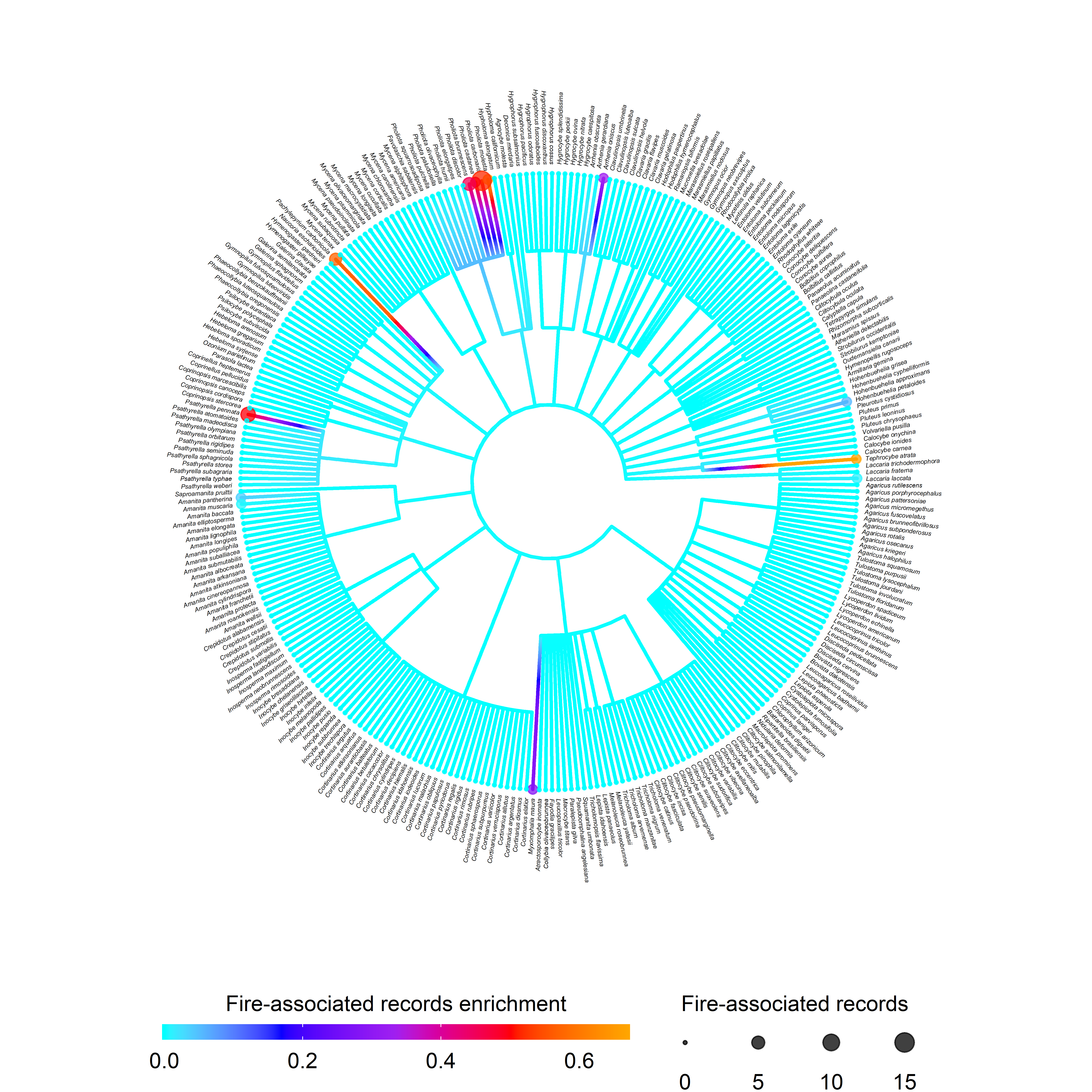
Figure 2.2: Circle cladogram showing fire-association enrichment for the top 300 Agaricales species with the highest enrichment