Chapter 3 Geographic analysis
Under construction…
3.1 Clean coordinates
Some occurrence records may have invalid coordinates (e.g., not in decimal format, not between -180 and 180 [lon] or -90 and 90 [lat], not within the bounds of the country listed) or coordinates with suspicious validity (e.g., zero/zero coordinates, lat and long are equal, coordinates match the country centroid), and these issues must be dealt with before conducting geographical analyses. coord_clean can be used to remove bad coordinates using a variety of validity tests (see function details). Additionally, the number of digits can be specified for automatic rounding of coordinates.
Example code for cleaning coordinates:
library(fungarium)
#import sample data set
data(agaricales_updated) #United States Agaricales records
#see Table 2.1 for preview of agaricales data set| id | institutionCode | scientificName | scientificNameAuthorship | recordedBy | year | eventDate | occurrenceRemarks | habitat | associatedTaxa | country | stateProvince | county | decimalLatitude | decimalLongitude | references | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 269864 | 9961160 | O | Galerina mniophila | (Lasch) Kühner | Gro Gulden | 1997 | 1997-11-15 | young Douglas fir forest | United States | Oregon | Benton | https://mycoportal.org/portal/collections/individual/index.php?occid=9961160 | ||||
| 313673 | 454144 | MICH | Mycena galopus | (Pers.) P. Kumm. | A. H. Smith | 1935 | 1935-11-30 | USA | California | Humboldt | 41.0595 | -124.1419 | https://mycoportal.org/portal/collections/individual/index.php?occid=454144 | |||
| 157092 | 4570238 | iNaturalist | Armillaria mellea | (Vahl) P. Kumm. | Cindy Trubovitz | 2017 | 2017-01-07 | <p>Growing from the base of an old log</p>; <a href=‘https://www.inaturalist.org/observations/5244247’ target=’_blank’ style=‘color: blue;’> Original observation #5244247 (iNaturalist)</a> | United States | California | 32.80975 | -117.1295 | https://mycoportal.org/portal/collections/individual/index.php?occid=4570238 | |||
| 281279 | 456630 | MICH | Hygrophorus olivaceoalbus | (Fr.) Fr. | R. L. Shaffer | 1956 | 1956-09-16 | USA | Idaho | Valley | 44.8753 | -115.9017 | https://mycoportal.org/portal/collections/individual/index.php?occid=456630 | |||
| 32558 | 2270252 | MICH | Cortinarius | (Pers.) Gray | S. J. Mazzer | 1970 | 1970-05-31 | [Collection is stored in Cortinarius indeterminate box] | USA | Michigan | Cheboygan | https://mycoportal.org/portal/collections/individual/index.php?occid=2270252 | ||||
| 522392 | 345316 | MICH | Crepidotus malachius var. phragmocystidiosus | Hesler & A.H. Sm. | A. H. Smith | 1959 | 1959-08-18 | Notes with collection North Amer. Sp. Crepidotus 1965 pg. 58. | On hardwood | USA | Michigan | Chippewa | https://mycoportal.org/portal/collections/individual/index.php?occid=345316 | |||
| 234305 | 526338 | OSC | Cortinarius montanus | Kauffman | Jim Trappe | 1976 | 1976-09-08 | Old-growth Tsme | Old-growth Tsme | USA | Oregon | Lane | https://mycoportal.org/portal/collections/individual/index.php?occid=526338 | |||
| 356189 | 2287069 | DUKE | Schizophyllum commune | Fr. | D. Porter | 1998 | 1998-04-00 | United States | Georgia | Clarke | 33.95 | -83.383333 | https://mycoportal.org/portal/collections/individual/index.php?occid=2287069 | |||
| 237907 | 2357852 | WTU | Cortinarius rubicundulus | (Rea) A. Pearson | L. L. Norvell, SAR. | 1992 | 1992-11-17 | Rotten wood/ duff. Sequoia, Tan Oak. | U.S.A. | California | Mendocino | https://mycoportal.org/portal/collections/individual/index.php?occid=2357852 | ||||
| 140361 | 6920708 | iNaturalist | Amanita pachycolea | D.E. Stuntz | Taylor Bates | 2017 | 2017-11-13 | <a href=‘https://www.inaturalist.org/observations/8797347’ target=’_blank’ style=‘color: blue;’> Original observation #8797347 (iNaturalist)</a> | United States | Oregon | 44.665631 | -123.242596 | https://mycoportal.org/portal/collections/individual/index.php?occid=6920708 |
#clean coordinates
agaricales_cleaned <- coord_clean(agaricales_updated) ## 'non-numeric coord' test: 11788 records removed.## 'non-valid coord' test: 0 records removed.## 'zero' test: 0 records removed.## 'equal' test: 0 records removed.## Running 'countries' test...## 'countries' test: 392 records removed.## Running 'centroids' test...## 'centroids' test: 0 records removed.## Total records removed: 121803.2 Assigning records to geographic grid cells
Before plotting geographic trait data within bounding “boxes,” record coordinates are used to assign each record to individual grid cells. hex_grid can be used to create a world map grid of hexagonal cells with a specific area (in square kilometers). See the code below for a demonstration on how to assign occurrence records to individual grid cells.
Example code for finding trait records and calculating enrichment factors:
#make hex grid
grid <- hex_grid(20000)
#convert lat/long points to sf geometry
agaricales_sf <- sf::st_as_sf(agaricales_cleaned,
coords = c("decimalLongitude", "decimalLatitude"),
crs = "+proj=latlong +ellps=WGS84 +datum=WGS84")
#convert to cylindrical equal area projection coordinates
agaricales_sf <- sf::st_transform(agaricales_sf,
crs = "+proj=cea +ellps=WGS84 +datum=WGS84")
#assign points to grid cells
agaricales_grid <- sf::st_join(grid, agaricales_sf, join=sf::st_contains, left=FALSE)3.3 Calculate trait enrichment per grid cell
After assigning records to specific grid cells, trait enrichment can be calculated for each cell using the enrichment function. See code below for an example on calculating fire-associated enrichment.
Example code for calculating enrichment factors:
#find fire-associated records
string1 <- "(?i)charred|burn(t|ed)|scorched|fire.?(killed|damaged|scarred)|killed.by.fire"
string2 <- "(?i)un.?burn(t|ed)"
trait_rec <- find_trait(agaricales_grid, pos_string=string1, neg_string=string2)
#find fire-associated enrichment per grid cell
fire_enrich <- enrichment(agaricales_grid, trait_rec, ext_var="geometry", by="hex")
#filter out hex cells with low amount of occurrences
fire_enrich <- fire_enrich[fire_enrich$freq>=5,]3.4 Visualize trait enrichment
Once enrichment values have been calculated, annotated grid cells can be plotted on a world map using trait_map. Additionally, world map can be cropped to a specific region, if desired.
Example code for making trait map:
#plot trait data on world map
library(ggplot2)
fire_enrich <- sf::st_as_sf(fire_enrich) #convert enrichment table back to sf
map <- trait_map(fire_enrich, aes_mapping = aes(fill=trait_ratio),
color=NA, alpha=0.95, size=0.1)
map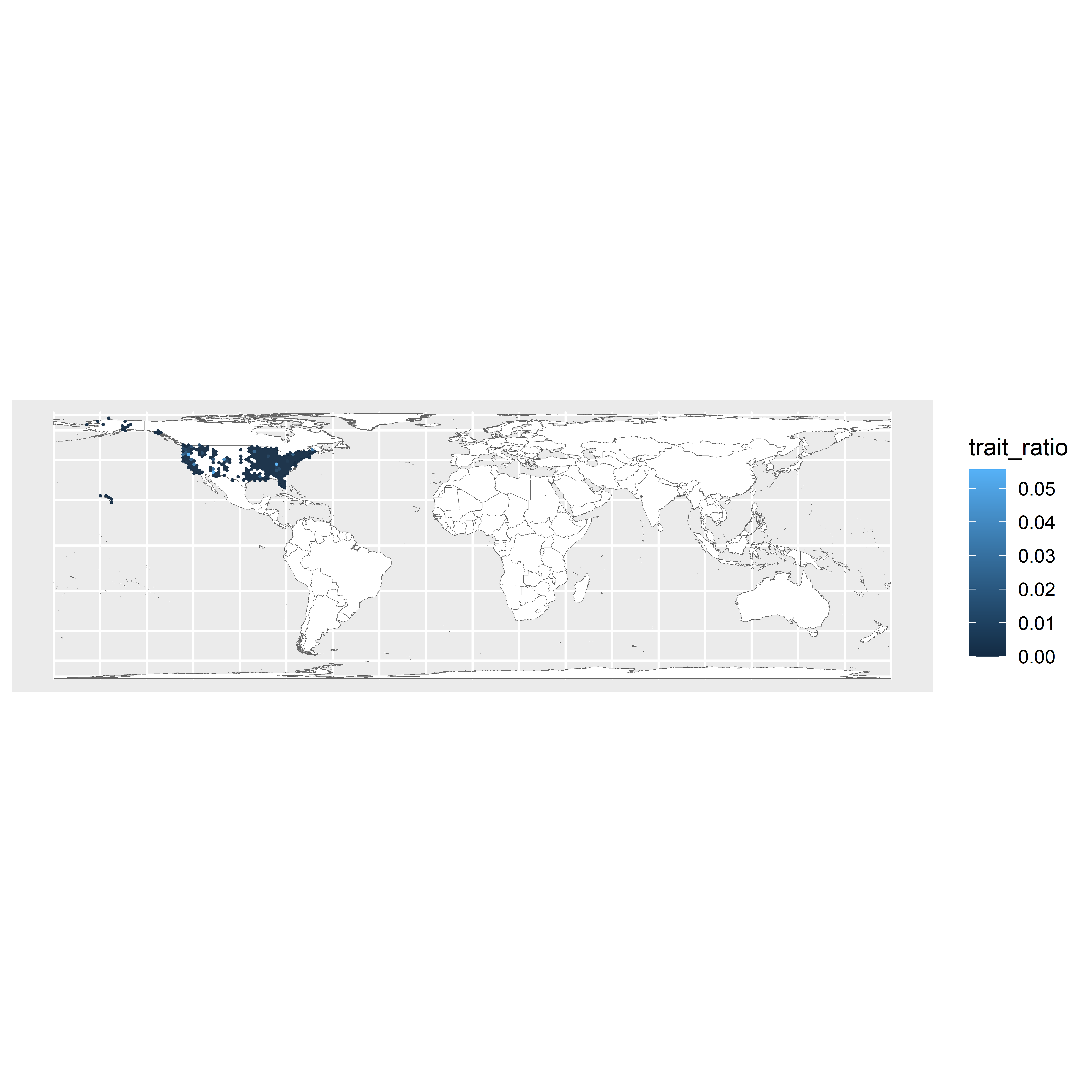
Figure 3.1: World map showing fire-associated enrichment via 20,000km2 grid cells
Example code for making trait map of specific region:
#Because sample data in from US only, crop world map to focus on US region
map+
ylim(c(300000,6000000))+
xlim(c(-18000000,-6000000))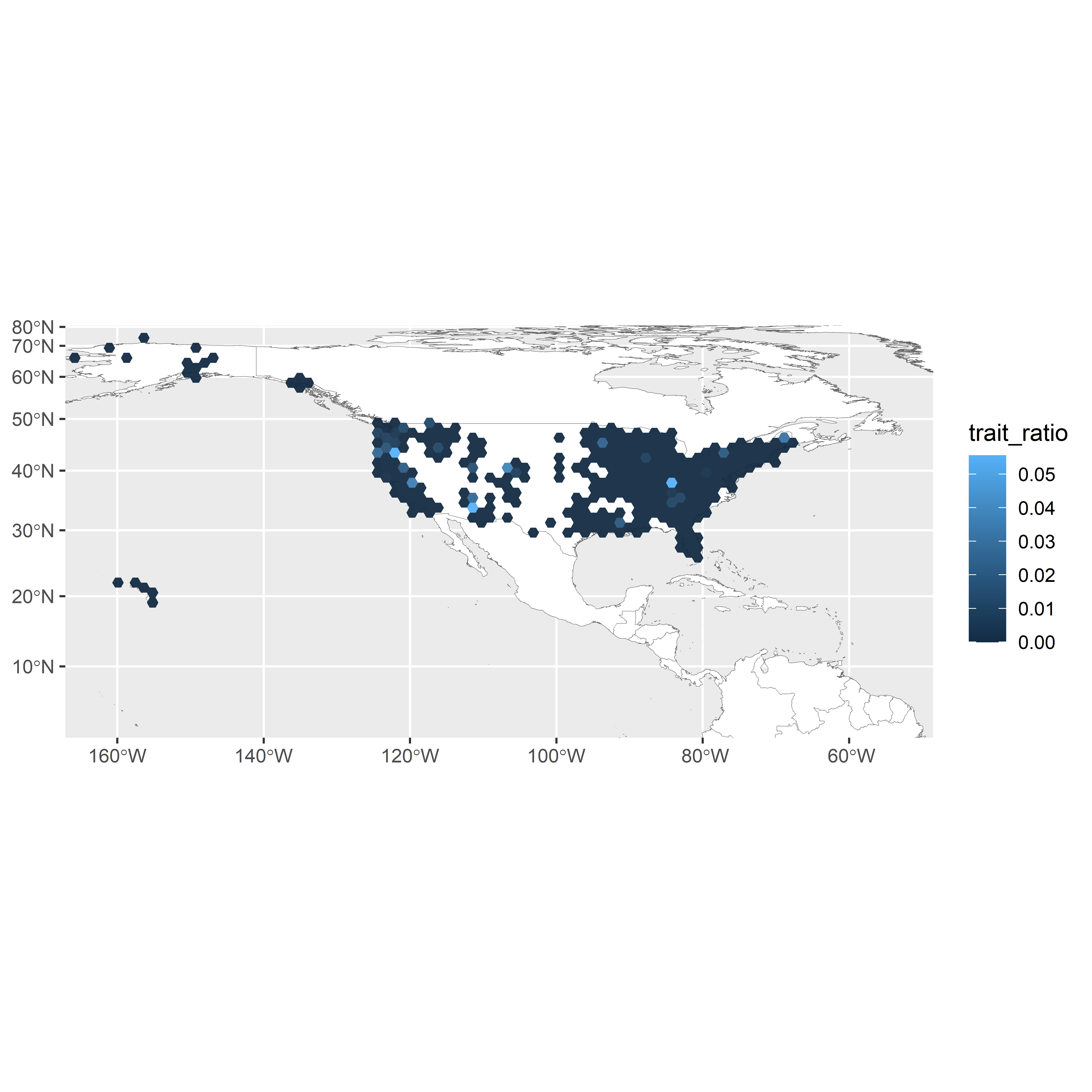
Figure 3.2: United States map showing fire-associated enrichment via 20,000km2 grid cells